Orojenesis
Orojenesis is an approach to compute data movement bounds for tensor algorithms. It comprehends reuse and the ability of a buffer to exploit reuse to reduce data movement and provides a bound that no dataflow or mapping can possibly exceed under varying on-chip buffer capacity constraints, including mappings that fuse a sequence of tensor operations to exploit producer-consumer reuse.
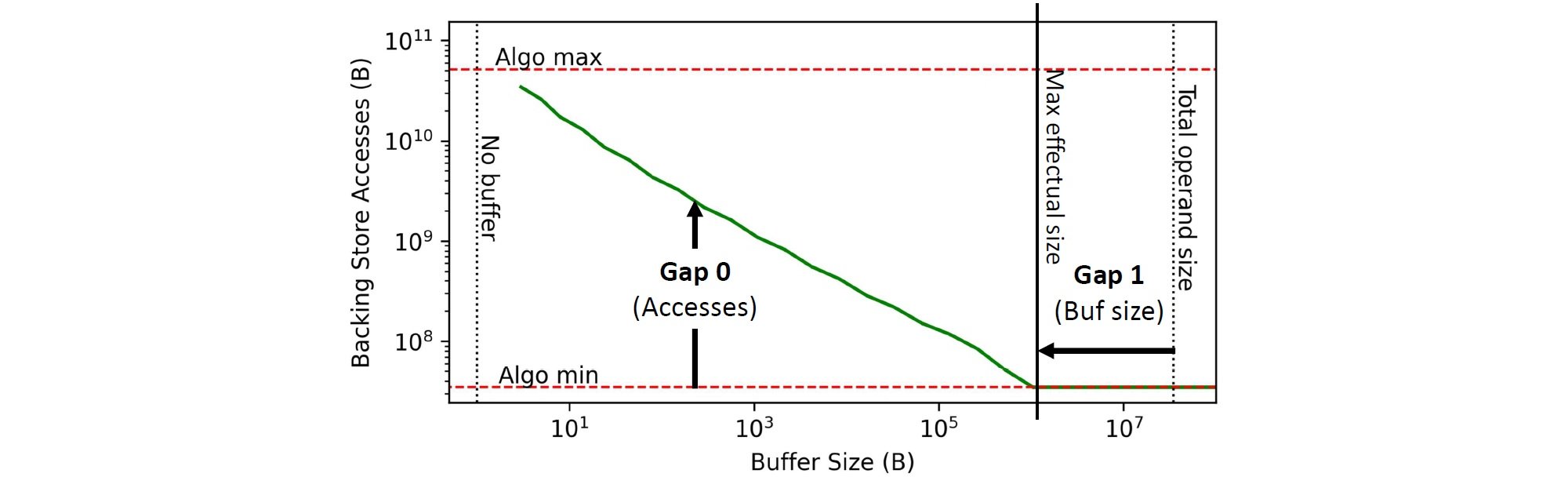
Orojenesis generates a "ski-slope diagram" that shows the relationship between a buffer’s size and the lower data movement limit to/from the next level in a memory hierarchy.
Ski-slope Diagram
For more details, please refer to:
@inproceedings{
huang2024isca,
title={Mind the Gap: Attainable Data Movement and Operational Intensity Bounds for Tensor Algorithms},
author={Qijing Huang and Po-An Tsai and Joel S Emer and Angshuman Parashar},
booktitle={International Symposium on Computer Architecture (ISCA)},
year={2024}
}1. Installation
First, download the Timeloop github and checkout the Orojenesis branch.
git clone --recurse-submodules -b oaves_keep_max https://github.com/NVlabs/timeloop.git
cd orojenesis1.1. Docker Setup [OPTIONAL]
If you don't have sudo access on your system, please consider using a Docker container.
A Dockerfile is provided with the project for setting up the software dependencies.
To build the container image, navigate to the root of the orojenesis repository and execute the following command:
docker build -f ./docker/Dockerfile -t orojenesis .To start the container, use the following command:
docker run -it orojenesis -p 8888:8888 -v $(pwd):/home/workspace bashOnce the docker is running, please follow the instructions to finish the installation and run the artifact.
1.2. Software Installation
Install software dependencies by running under the orojenesis directory:
./install.sh2. Run Orojenesis
Orojenesis take Einsums as input and produces the corresponding ski-slope diagrams. This section demonstrates how to customize workload definitions and mapper constraints for Orojenesis bound generation.
-
Workload Definition: The workload definition describes the tensor workload being analyzed.
- Predefined Workload Classes:
We provide a base class named Op in
src/utils.pythat serves as an abstraction for different workload types. Currently, it supports convolution (Conv) and grouped batched matrix multiplication (GBMM). - Defining New Einsum Shapes: If you need to handle a new Einsum shape beyond Conv and GBMM, you can easily extend the functionality by following the template provided in the Op class.
- Problem Definition Output:
The
to_yamlfunction is responsible for converting the workload definition into a YAML format that adheres to the Timeloop problem format.
- Predefined Workload Classes:
We provide a base class named Op in
-
[Optional] Mapper: The mapper specifies the search strategy and mapping constraints.
- Generic Mapper: We provide a generic mapper in
configs/single-einsum/mapper.yamlthat can work for most Einsum shapes. - Workload-Specific Constraints: If you have knowledge of suboptimal or irrelevant search space options specific to your workload, you can define additional constraints in the mapper_constraints section of the mapper file. An example of this is provided in
configs/single-einsum/conv_mapper.yaml.For Conv workloads. For more details on Timeloop mapper constraints,, please refer to Timeloop mapper constraints.
- Generic Mapper: We provide a generic mapper in
The Snowcat architecture is defined in ./outputs/single-einsum/arch.yaml. In most cases, you won't need to modify this file for an single Einsum.
2.1 Example for generating the bound for 1x1 Convolution
First we need to import the orojenesis utility functions and set the TIMELOOP_BASE_PATH to point the root directory of Timeloop.
import os
if "TIMELOOP_BASE_PATH" not in os.environ:
timeloop_path = input("Please specify the path to Timeloop repo (default: " + os.getcwd() + "/../):" ) or os.getcwd() + "/../"
os.environ["TIMELOOP_BASE_PATH"] = timeloop_path
os.environ["TIMELOOP_DIR"] = timeloop_path
os.environ["TIMELOOP_ENABLE_FIRST_READ_ELISION"] = "1"
print("Path to timeloop repo: ", os.environ["TIMELOOP_BASE_PATH"])
import pathlib
import src.utils as utilsLet's assume we want to derive Orojenesis bounds for a 1x1 convolution with input channel size 32 and output channel size 16. Here's how to define the problem using the Conv class:
# Define the workload shape.
prob = utils.Conv(R=1, S=1, C=32, K=16)
mapper_yaml = pathlib.Path('./configs/single-einsum/conv_mapper.yaml')
# Specify output directory
output_dir = pathlib.Path('./outputs/single-einsum')
arch_yaml = pathlib.Path('./configs/single-einsum/arch.yaml')
utils.GenerateBound(prob, output_dir, arch_yaml, mapper_yaml, keep_one_best_entry_across_buf=True)
# Output CSV paths
stats_files = utils.get_stats_files(output_dir, [prob])
print(f'Output CSV file: {stats_files[0]}')Interpreting the CSV output:
- Column 0: the buffer size in ascending order
- Column 1: the corresponding achievable operational intensity (OI)
- Column 2: the corresponding achievable DRAM access count
- Column 3: the mapping shortform
3. More Examples
We provide Jupyter notebooks, orojenesis/orojenesis_single.ipynb
and orojenesis/orojenesis_multi.ipynb, to guide you through generating
the key examples in the paper. Please launch the Jupyter GUI
under orojenesis by running:
jupyter notebookIf a GUI is not accessible, you can convert the notebook into Python scripts by using:
jupyter nbconvert --to script <my-notebook.ipynb>and launch the Python script
python <my-notebook.py>The output figures will be saved to orojenesis/figs folder.